How I am trying to find something new about biliary atresia using cutting-edge techniques
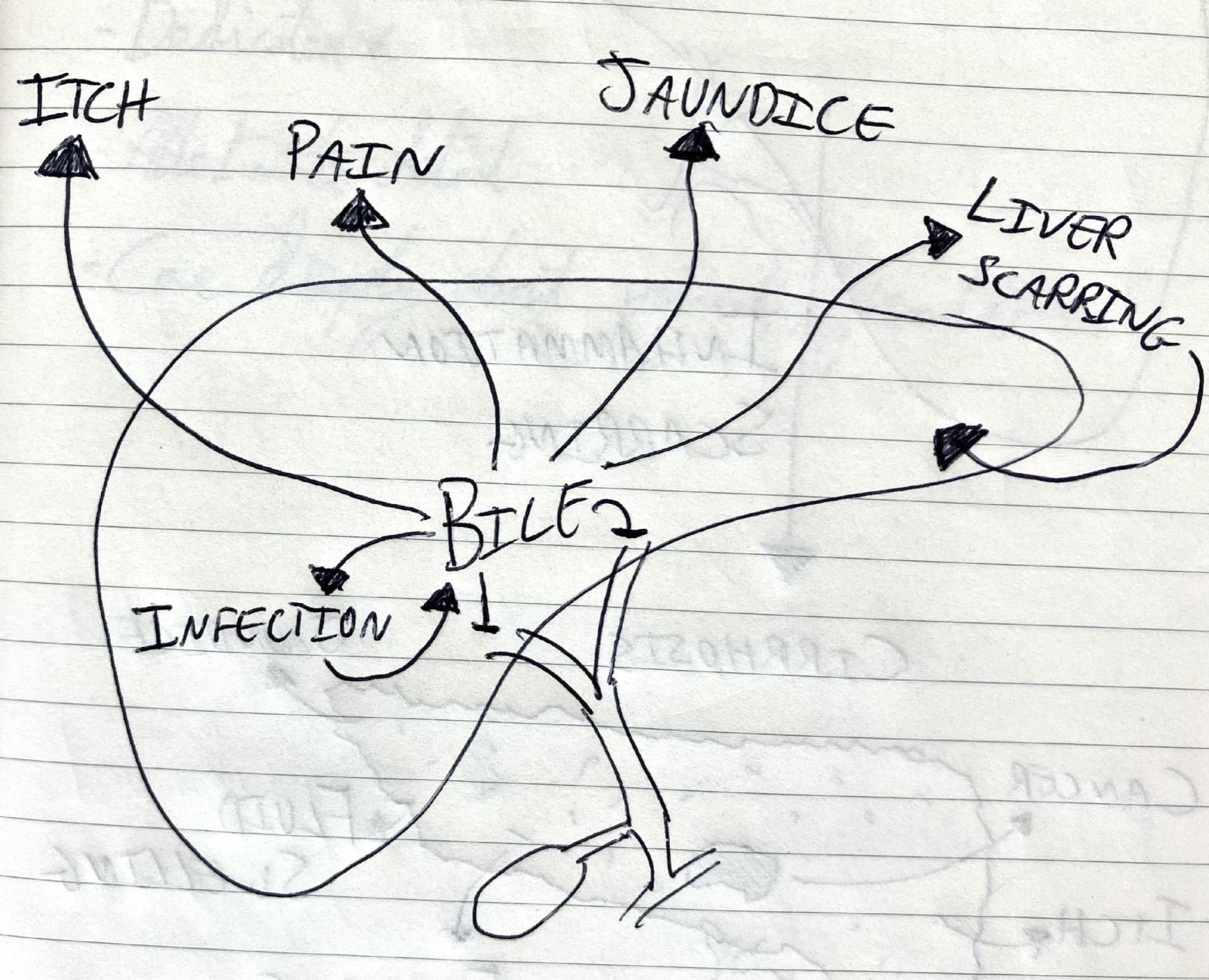
Over the last couple of years, I’ve started working on biliary atresia. This is a rare condition where the bile ducts, which normally drain the liver, become scarred and disappear within the first few months of life. Whilst it is rare (affecting fewer than 100 children in the UK each year), it is the most common reason a child needs a liver transplant. This is the main reason I decided to move into this field.
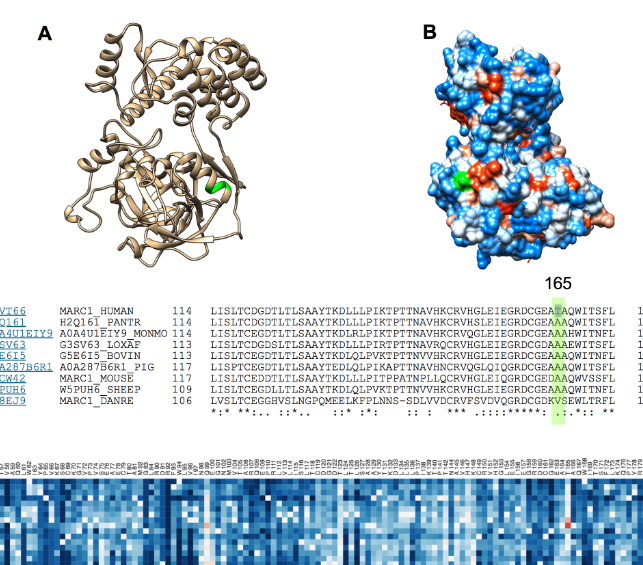
However, it’s tricky to know where to start when beginning in a near area. Many very smart people have been working hard on this condition for a long time. So, I decided to try out some brand-new techniques that allow you to study the liver (or anything else) one cell at a time. The technical name for this is single-cell spatial transcriptomics. This means we can take a piece of tissue—for example, a liver biopsy—and see what genes each cell is making. This tells us what each cell is doing and helps identify what kind of cell it is.
I’ve been fortunate to receive funding from Guts-BSPGHAN, ESPGHAN, and the Royal Society to explore aspects of this research, in addition to my ongoing funding from the NIHR.

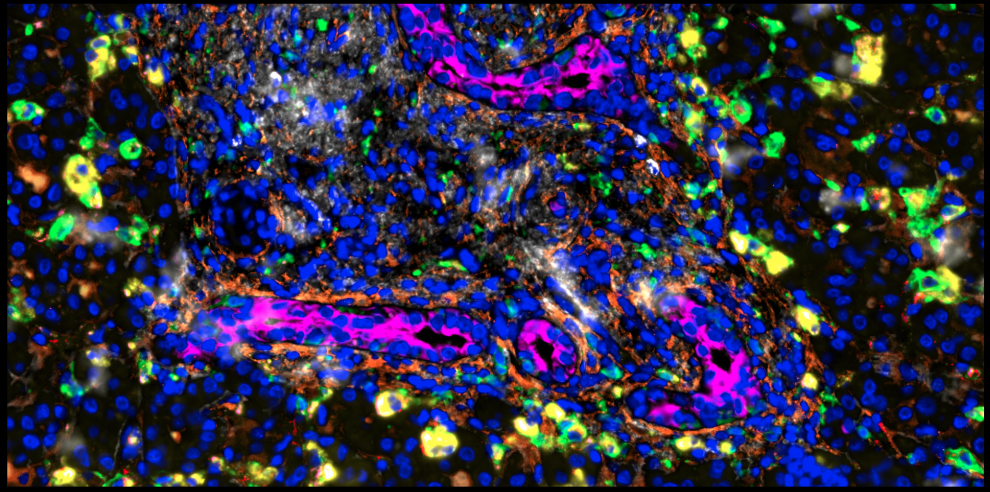
Whilst the full results aren’t ready for publication yet, the images are fascinating. The picture shows some liver tissue taken during the operation for biliary atresia (at around two months old). In brown are the main liver cells (‘hepatocytes’), in blue are the scar-forming cells (‘fibroblasts’), in yellow are the bile duct cells (‘cholangiocytes’), and everything else is a combination of white blood cells and blood vessel cells.
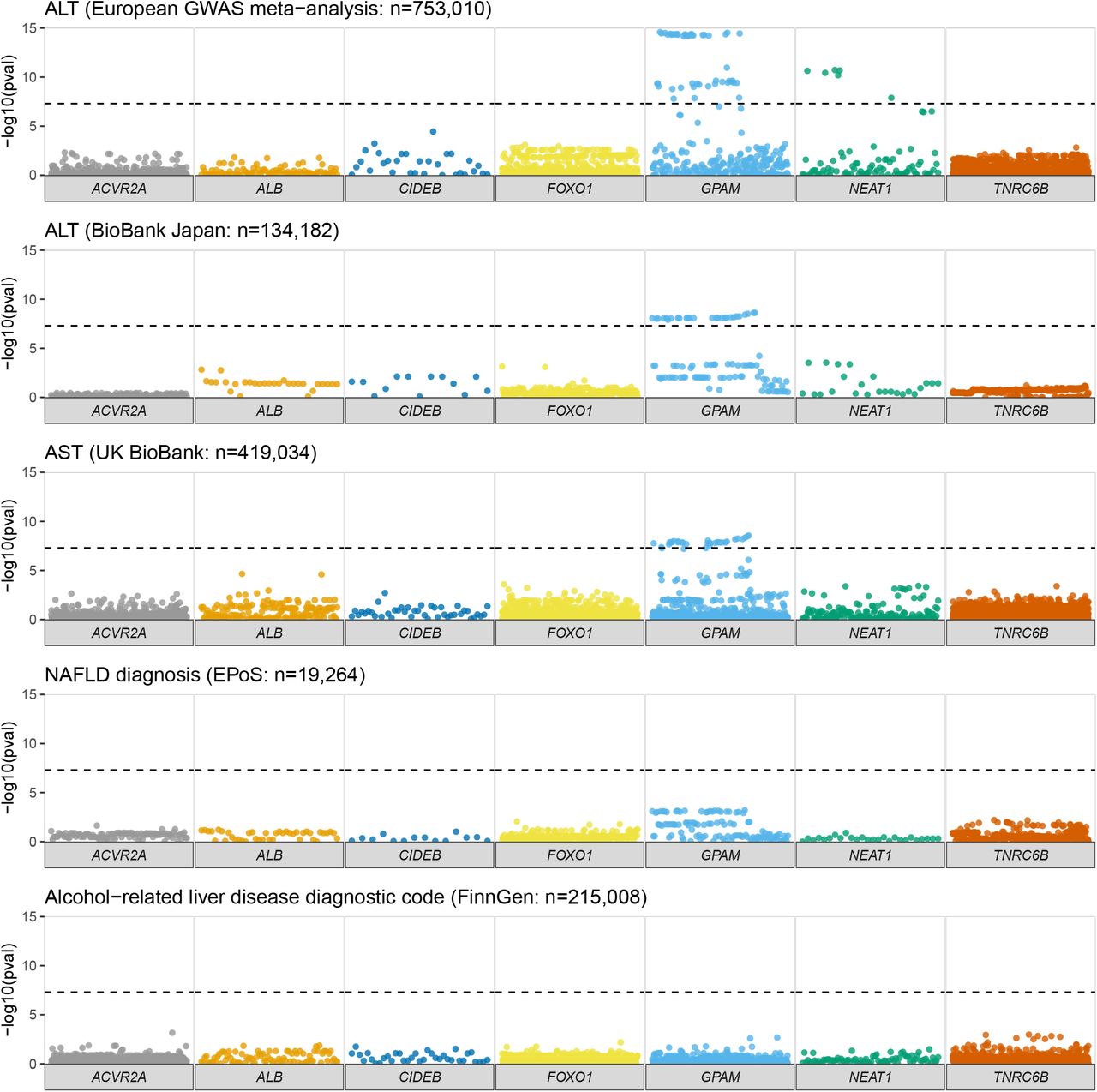
What’s great about this kind of research is that it generates new ideas for further work (‘hypothesis-generating’). Using advanced statistics and mathematics, it’s possible to predict how cells are communicating with each other.
What I’m doing now is conducting more experiments to check that what I’ve found so far is true when tested with other methods. Each approach has its advantages and disadvantages, and we’ve already learned a lot about what this technique (‘CosMx’) can achieve and where we need to be careful to extract the right information.
I’m hopeful that I’ll be able to share the full story of what I’ve discovered in the next year or so. In the meantime, please get in touch if you’d like to discuss CosMx or single-cell spatial transcriptomics. I’m always eager to collaborate with new people, and there are opportunities to join me here in Birmingham.